Genetic diversity and intraspecific mitochondrial DNA variations in the Georgian Mountain breed of Bos taurus reveal admixture, introgression and potential parallel vs. convergent evolution patterns
Abstract
This study elucidates the haplotype diversity and mechanisms of evolutionary divergence for a broad population of the Georgian Mountain breed (GMB) of Bos taurus, using the sequencing and analysis of its mitochondrial DNA (mtDNA). In the evolutionary analyses, sequences of the targeted mtDNA region, involving the D-loop, CYTB, tRNA-Thr, and tRNA-Pro encoding genetic loci were analyzed using MEGA11, DnaSP, and SplitsTree software packages. A total of 25 haplotypes were determined among 82 individuals of GMB, belonging predominantly to the haplogroups T (T3, T1, T2, T4) or Q (Q1). Ten singleton haplotypes could also be determined in the GMB population. In the maximum likelihood evolutionary analysis, the singleton haplotype SNGT-9 appeared to be most closely related to the Bos indicus sub-haplogroup I1a. The haplotype diversity (0.997), nucleotide diversity (0.00636) and the overall mean distance within a population (0.01) calculated for GMB were greater as compared to the respective estimates (0.930, 0.00482 and 0.00) determined for its closest cattle relatives globally, suggesting stronger selection. It is suggested that the GMB diversity has been shaped by both parallel and convergent evolution, as well as by possible introgression, while pinpointing this breed’s ancient origin collectively.
Keywords
Georgian Mountain breed, cattle, haplotype, haplogroup, genetic diversity, population structure, mtDNA
Introduction
Climate change, driven by global warming, threatens agricultural animals' survival in the Anthropocene (Elayadeth-Meethal et al., 2018). Smaller body size in mammals is thought to aid adaptation to warmer climates (Pacifici et al., 2017), as there appears to be an interplay between body size reduction and increased livestock tolerance to warming (Elayadeth-Meethal et al., 2018). In this light, preserving biodiversity, conserving endemic cattle breeds of small body size, and deciphering their population structures and evolutionary mechanisms are important, given global food security pressures and climate change (Mitchell et al., 2018).
According to the archeozoological and genetic evidence, mainly relying on the results of mitochondrial DNA (mtDNA) analyses, modern cattle are thought to have emerged via just two independent and geographically distinct domestication events of aurochs (Bos primigenius), both of which occurred in southwest Asia: one event is thought to have occurred in the Fertile Crescent, resulting further into modern taurine breeds of B. taurus, while the other event took place in the Indus Valley, leading to the emergence of modern zebuine breeds of its subspecies B. indicus (Achilli, Olivieri, & Pellecchia, 2008). The diversity of modern cattle has been structured largely into three major groups represented by Eurasian taurine, African taurine and Asian indicine cattle; these include different types of crosses, all combinations and intricacies (Kim, 2020), with various breeds exhibiting predominantly the macro-haplogroup T of B. taurus (Achilli et al., 2008).
While the genetic diversity of many cattle breeds from the above groups has been well characterized, there remains a significant lack of information specifically concerning the Georgian Mountain breed (GMB) of B. taurus endemic to the Caucasus region. GMB individuals are very small in body size, with the live weight varying from 220 to 280kg for mature cows, and from 270 to 370kg for bulls (Kunelauri, Gogniashvili, Tabidze, Basiladze, & Beridze, 2019). The coat colours of GMB individuals are black, black-and-white or red-and-white. Compared with other breeds from this region, GMB not only demonstrates stronger endurance and enhanced sustainability towards the above conditions but is also less susceptible to impoverished food (Kunelauri et al., 2019). This breed has been well adapted to the harsh climate and other conditions of the Caucasus mountains, and to its grazing lands with slopes that sometimes reach an angle of 45 degrees (Kunelauri et al., 2019). Two initial studies by (Kunelauri et al., 2022; Kunelauri et al., 2019) analyzing the sequences of mtDNA loci versus a mitogenome in a very limited number of cattle individuals, revealed B. taurus haplogroups T and Q (branch Q1) and suggested the presence of some unknown haplotypes as well in this breed.
Our study aimed to characterize the haplotypes across broad GMB populations from the Khevsureti and Adjara regions of Georgia, and to gain initial insights into their evolutionary mechanisms by analyzing mtDNA genetic loci.
Materials and methods
Animal sampling
A total of 82 GMB individuals were sampled across the Khevsureti and Adjara regions of Georgia, as these regions have a high concentration of this breed, in 2019–2022, selecting unrelated animals based on pedigree information to minimize kinship. Only one individual was selected and sampled per village across the above regions. Specifically, hair follicle samples from the selected Khevsurian (n = 36) and Adjarian (n = 46) cattle were obtained for mtDNA extraction, PCR amplification, and sequencing.
Primer design, PCR amplification and sequencing of targeted mtDNA
The Tissue and Hair Extraction Kit, coupled with the DNA IQ™ kit (Promega, Inc., Madison, WI, USA), and the Quick-DNA Microprep Plus Kit (Zymo Research, Inc., Irvine, CA, USA), were used for the extraction of mtDNAs from the hair follicle samples. The PrimerQuest™ Tool (Integrated DNA Technologies, USA) was used to design primers for PCR amplification and sequencing of the targeted mitochondrial genome region. The designed forward and reverse primers (5'-CCAACAAACTAGGAGGAGTA-3' and 5'-CGCGGCATGGTAATTAAG-3') allowed us to amplify the 810-bp mtDNA region encompassing the genes encoding for tRNA-Thr, tRNA-Pro, as well as the CYTB gene and the D-loop loci across the above mitochondrial region from the selected GMB individuals. PCR conditions were 94°C for 5min, followed by 35 amplification cycles, each consisting of sequential incubation at 94°C (30s), 51°C (30s), and 72°C (1min and 20s), with the final 72°C (5min) extension.
The DNA sequencing of the PCR-amplified products was performed in both directions, using the BigDye Terminator v3.1. cycle sequencing kit (Applied Biosystems, Inc., Foster City, CA or Thermo Fisher Scientific) or the BrilliantDye™ Terminator (v3.1) Cycle Sequencing Kit (NimaGen, Nijmegen, the Netherlands). The post-cycle sequencing reaction contaminants were removed by applying the ZR DNA Sequencing Clean-Up Kit (Zymo Research, Inc., Irvine, CA, USA). The 3100xl Genetic Analyzer (Applied Biosystems, Inc., Foster City, CA, USA) was used to separate the labelled DNA fragments by size; the Geneious Prime v. 7.0.9 (Biomatters, Inc., Boston, MA, USA) and Sequencher v. 5.4.6 (Gene Codes, Corp. Ann Arbor, MI, USA) were utilized to edit and assemble the consensus sequences. All low-quality sequences were trimmed from both forward and reverse DNA sequence reads. The representative mtDNA sequences, obtained from the above DNA sequencing experiments, were submitted to, and are available under the accession numbers OR412787-OR412811, the GenBank database of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/).
Evolutionary analyses
The MEGA11 (v. 11.0.13) and DnaSP (v. 6.12.03) software packages were used to determine the population structures and evolutionary features of GMB individuals and their genetically closest cattle from different breeds of B. taurus and B. indicus. For these analyses, the respective mtDNA regions of 77 genetically closest cattle (Supplemental Table 1) were selected in the NCBI nucleotide database, using megaBLAST with the default parameters (Expected threshold - 10, Word size - 28, Match/mismatch scores - 1,-2, Gap costs - Linear, Extension - 2). The evolutionary analysis included representatives of each haplotype from GMB and its genetically closest cattle individuals preselected based on DNA identity, query coverage and E values from the BLAST analysis. Specifically, for evolutionary analyses, we selected the 491-bp mtDNA region, encompassing the CYTB and D-loop loci, as well as the genes encoding for tRNA-Thr and tRNA-Pro, which were shared exclusively by all these organisms. In this subset, we performed a random selection of the breed representatives from their country of origin, when they exhibited the BLAST-generated multiple identical hits to the query DNA sequences in the NCBI GenBank database. MEGA11 was used to determine specifically the shape parameter for the discrete gamma distribution to model evolutionary rates, as well as nucleotide substitution patterns and rates across the targeted mtDNA genetic loci as recommended previously (Tamura & Nei, 1993). In the MEGA analyses, Maximum Likelihood (ML) estimates of the transition/transversion bias were also determined, by applying the Kimura 2-parameter model (Kimura, 1980). Maximum Composite Likelihood (MCL) estimates of the pattern of nucleotide substitution were also determined as described by (Tamura, Nei, & Kumar, 2004). The ML method and the Tamura-Nei model (Tamura & Nei, 1993) were used to infer the evolutionary relationships between the GMB individuals and their genetically closest cattle individuals.
The haplotypes and their diversity, as well as polymorphic (segregating) sites, nucleotide diversity (Pi), invariable (monomorphic) versus variable (polymorphic) sites, and singleton variable versus parsimony informative sites across the 491-bp mtDNA region were determined using DnaSP. The haplogroup was determined for each GMB individual according to the haplogroup classification described previously (Xia, 2021). The degree of linkage disequilibrium (LD) (including the ZZ-value) was assessed as recommended by (Rozas, Gullaud, Blandin, & Aguadé, 2001). As the standardized measure of LD, the ZZ-value was derived from the squared correlation coefficient (r²) between pairs of polymorphic sites, and the variance of the r² values across multiple loci.
The two-tailed Fisher's (F) exact test and the chi-square (Chi-sq) test were applied to determine whether the associations between polymorphic sites were significant, with Bonferroni correction additionally employed for these tests as implemented in DnaSP.
In the genetic recombination analysis, a minimum number of putative recombination events (Rm), and the coordinates of recombination hotspots were determined when detected by DnaSP across the targeted mtDNA regions of the cattle individuals. For re-examining recombination inferences, we also applied the SplitsTree (version 4.14.4) and RDP4 (Beta 4.96) software packages. Particularly, we used the method of split decomposition method, implemented in SplitsTree, to detect parallel nucleotide substitutions with conflicting evolutionary signals in the targeted mtDNA region in the targeted cattle populations. Every subset of the mtDNA sequences that encompassed such parallel changes conjointly displayed across a SplitsTree-generated parallelogram(s) was subjected to the Phi (Pairwise Homoplasy Index) test (Bruen, Philippe, & Bryant, 2006) for measuring homoplasy. In the RDP4 analyses, searching for genetic recombination events, the RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, and 3Seq recombination detection algorithms were employed. We used the stringent approach: the predetermined Bonferroni-corrected P-values only in a range of ≤ 0.05 were considered statistically significant for the significant breakpoint clusters (99%) if detected in these analyses.
Results
DNA sequence polymorphism and phylogenetic analyses
Based on sequencing of a 810bp sequence of mtDNA a total of 25 haplotypes were determined among 82 GMB individuals, using DnaSP. The haplotypes, haplogroups, genetic sub-lineages/clusters, and geographic distribution of these cattle individuals are presented in Table 1.
As shown, a great majority of the GMB individuals belonged to the haplogroup T3 (42.6%) followed by Q1 (12.1%), T1 (3.6%), T2 (15.8%) and T4 (1.2%), as well as ten unique SNGTs not associated with any previously described haplotype of B. taurus.
In Figure 1, we display the ML tree constructed using the nucleotide analysis of the 491-bp mtDNA region, elucidating the population structures of the GMB haplotypes, their genetically closest cattle individuals of B. taurus worldwide, and their genetic relationships.
The ML analysis resulted primarily in the distribution of the GMB haplotypes, including several SNGTs, and their genetically closest cattle individuals across 13 major haplogroup-specific genetic clusters I-XIII (Figure 1 and Table 1). As shown, the mtDNA sequence profiles of specific GMB haplotypes represented by individuals 6K, 7K, 15K and 31A were identical to those of various breeds from the European, African and Southeast Asian regions, belonging to T3 haplogroup with T3119 included. The phylogenetic inferences, for the GMB singletons, exhibited their broad scattering patterns across the ML tree, demonstrating their close genetic relationships with different breeds from different cattle groups globally. These groups included collectively the Turano-Mongolian, Central European, Podolic, European dairy, Balkan, British Mountain, South Asian and Shorthorned Zebu (Bos indicus) cattle respectively from haplogroups T1-T4, Q, and the specific sub-lineage I1a of the lineage I1. It is important to indicate that while many GMB haplotypes could not be distinguished globally from various B. taurus breeds within the T and Q sub-lineages, certain SNGTs of the above Georgian breed (e.g. SNGT-1, SNGT-4, SNGT-7, SNGT-9, SNGT-10) either clustered separately within or fell outside these sub-lineages (Figure 1).
|
No. |
GMB ID |
Region |
GenBank ID |
Haplotype |
Haplogroup |
Major Genetic Cluster |
|---|---|---|---|---|---|---|
|
1 |
33KSh |
Khevsureti |
OR412798 |
T1-1 |
T1 |
VI |
|
2 |
18K |
Khevsureti |
OR412803 |
T1-2 |
T1 |
IX |
|
3 |
36KSh |
Khevsureti |
- |
T1-2 |
T1 |
IX |
|
4 |
13A |
Adjara |
OR412802 |
T2-1 |
T2 |
IX |
|
5 |
12A |
Adjara |
- |
T2-1 |
T2 |
IX |
|
6 |
37A |
Adjara |
OR412800 |
T2-2 |
T2 |
VIII |
|
7 |
23A |
Adjara |
OR412811 |
T2-3 |
T2 |
XIII |
|
8 |
35KSh |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
9 |
29KSh |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
10 |
28KSh |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
11 |
26KSh |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
12 |
36A |
Adjara |
- |
T2-3 |
T2 |
XIII |
|
13 |
32A |
Adjara |
- |
T2-3 |
T2 |
XIII |
|
14 |
14A |
Adjara |
- |
T2-3 |
T2 |
XIII |
|
15 |
22K |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
16 |
11K |
Khevsureti |
- |
T2-3 |
T2 |
XIII |
|
17 |
15K |
Khevsureti |
OR412808 |
T3-1 |
T3 |
XII |
|
18 |
13K |
Khevsureti |
- |
T3-1 |
T3 |
XII |
|
19 |
39AB |
Adjara |
- |
T3-1 |
T3 |
XII |
|
20 |
1A |
Adjara |
- |
T3-1 |
T3 |
XII |
|
21 |
7K |
Khevsureti |
OR412806 |
T3-2 |
T3 |
XI |
|
22 |
31KSh |
Khevsureti |
- |
T3-2 |
T3 |
XI |
|
23 |
6K |
Khevsureti |
OR412794 |
T3119-1 |
T3119 |
III |
|
24 |
31A |
Adjara |
OR412795 |
T3119-2 |
T3119 |
IV |
|
25 |
20A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
26 |
10A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
27 |
5A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
28 |
14K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
29 |
48AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
30 |
44AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
31 |
43AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
32 |
47AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
33 |
12K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
34 |
2A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
35 |
9A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
36 |
18A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
37 |
28A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
38 |
40AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
39 |
46AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
40 |
10K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
41 |
21K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
42 |
8A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
43 |
17A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
44 |
27A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
45 |
45AB |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
46 |
1K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
47 |
17K |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
48 |
6A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
49 |
16A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
50 |
26A |
Adjara |
- |
T3119-2 |
T3119 |
IV |
|
51 |
34KSh |
Khevsureti |
- |
T3119-2 |
T3119 |
IV |
|
52 |
24A |
Adjara |
OR412796 |
T4-1 |
T4 |
V |
|
53 |
2K |
Khevsureti |
OR412788 |
Q1-1 |
Q1 |
I |
|
54 |
8K |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
55 |
4K |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
56 |
19K |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
57 |
20K |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
58 |
15A |
Adjara |
- |
Q1-1 |
Q1 |
I |
|
59 |
22A |
Adjara |
- |
Q1-1 |
Q1 |
I |
|
60 |
29A |
Adjara |
- |
Q1-1 |
Q1 |
I |
|
61 |
24KSh |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
62 |
27KSh |
Khevsureti |
- |
Q1-1 |
Q1 |
I |
|
63 |
3A |
Adjara |
OR412789 |
QI-3 |
QNSG* |
I |
|
64 |
41AB |
Adjara |
- |
QI-3 |
QNSG* |
I |
|
65 |
23Ksh |
Khevsureti |
- |
QI-3 |
QNSG* |
I |
|
66 |
21A |
Adjara |
OR412791 |
II-1 |
N/D |
II |
|
67 |
19A |
Adjara |
- |
II-1 |
N/D |
II |
|
68 |
38A |
Adjara |
- |
II-2 |
N/D |
II |
|
69 |
30A |
Adjara |
OR412792 |
II-2 |
N/D |
II |
|
70 |
42AB |
Adjara |
OR412805 |
T3-3 |
T3 |
XI |
|
71 |
4A |
Adjara |
- |
T3-3 |
T3 |
XI |
|
72 |
34A |
Adjara |
- |
T3-3 |
T3 |
XI |
|
73 |
3K |
Khevsureti |
OR412787 |
SNGT-1 |
Q |
I |
|
74 |
25A |
Adjara |
OR412801 |
SNGT-2 |
T |
VIII |
|
75 |
30KSh |
Khevsureti |
OR412810 |
SNGT-3 |
T2 |
XIII |
|
76 |
35A |
Adjara |
OR412790 |
SNGT-4 |
N/D |
N/D |
|
77 |
33A |
Adjara |
OR412793 |
SNGT-5 |
N/D |
N/D |
|
78 |
25Ksh |
Khevsureti |
OR412797 |
SNGT-6 |
T1 |
VI |
|
79 |
9K |
Khevsureti |
OR412799 |
SNGT-7 |
T3 |
VII |
|
80 |
5K |
Khevsureti |
OR412807 |
SNGT-8 |
T3 |
XII |
|
81 |
16K |
Khevsureti |
OR412804 |
SNGT-9 |
N/D |
X |
|
82 |
32KSh |
Khevsureti |
OR412809 |
SNGT-10 |
T2 |
XIII |
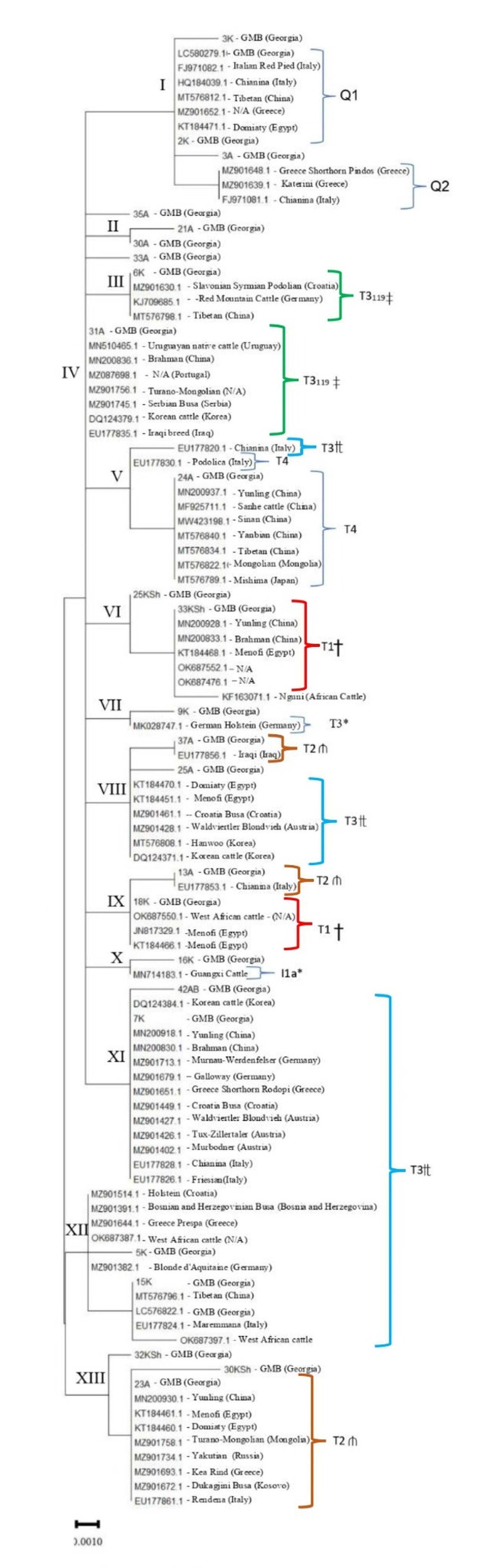
Moreover, according to the ML analysis, certain genetic groups from the same sub-haplogroups of T (except T4), determined previously (Xia, 2021), fell into two or more distantly related phylogenetic clades.
In Table 2, we describe polymorphisms of the targeted mtDNA region of the GMB haplotypes exhibiting previously unknown mutations within the B. taurus global populations.
|
Haplotype |
Allele frequency |
Polymorphisms and their coordinates across the targeted mtDNA region |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
15789 |
15813 |
15846 |
15878 |
15917 |
15919 |
15954 |
15961 |
15966 |
16055 |
16057 |
|
T3-3 |
3 |
C |
T |
T |
C |
A |
G |
G |
G |
G |
T |
G |
|
II-1 |
2 |
C |
C |
T |
C |
G |
A |
G |
G |
G |
T |
G |
|
II-2 |
2 |
C |
T |
T |
C |
G |
A |
G |
G |
G |
T |
G |
|
QI-1 |
13 |
C |
T |
T |
C |
A |
A |
G |
A |
G |
T |
G |
|
SNGT-1 |
1 |
C |
T |
T |
C |
A |
A |
G |
G |
G |
C |
G |
|
SNGT-2 |
1 |
T |
T |
T |
C |
A |
A |
G |
G |
G |
T |
G |
|
SNGT-3 |
1 |
C |
T |
T |
C |
A |
A |
G |
G |
A |
T |
C |
|
SNGT-4 |
2 |
C |
T |
C |
C |
A |
A |
G |
G |
G |
T |
G |
|
SNGT-5 |
1 |
C |
T |
T |
C |
A |
A |
A |
G |
G |
T |
G |
|
SNGT-8 |
1 |
C |
T |
T |
T |
A |
A |
G |
G |
G |
T |
A |
|
SNGT-10 |
1 |
C |
T |
T |
C |
A |
A |
G |
G |
G |
T |
T |
Specifically, a total of 11 unique DNA polymorphisms could be identified in this subset, being mainly G↔A and T↔C nucleotide substitutions. In addition, Table 3 displays the ML estimates of the substitution matrix, as well as the MCL estimates of the pattern of nucleotide substitution.
|
Targeted population |
|
ML estimate of substitution matrix |
MCL estimate of the pattern of nucleotide substitution |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
A |
T/U |
C |
G |
|
A |
T/U |
C |
G |
|
GMB |
|
|
|
|
|
|
|
|
|
|
|
A |
- |
1.13 |
0.98 |
10.5 |
|
- |
2.07 |
1.79 |
10.14 |
|
|
T/U |
1.83 |
- |
22.25 |
0.56 |
|
3.36 |
- |
18.6 |
1.02 |
|
|
C |
1.83 |
25.66 |
- |
0.56 |
|
3.36 |
21.45 |
- |
1.02 |
|
|
G |
33.05 |
1.13 |
0.98 |
- |
|
33.34 |
2.07 |
1.79 |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
GMB closest cattle relatives globally |
|
|
|
|
|
|
|
|
|
|
|
A |
- |
2.20 |
1.90 |
9.37 |
|
- |
1.97 |
1.71 |
9.57 |
|
|
T/U |
3.57 |
- |
19.59 |
1.08 |
|
3.21 |
- |
20.03 |
0.97 |
|
|
C |
3.57 |
22.62 |
- |
1.08 |
|
3.21 |
23.13 |
- |
0.97 |
|
|
G |
30.90 |
2.20 |
1.90 |
- |
|
31.56 |
1.97 |
1.71 |
- |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Both cattle groups combined |
|
|
|
|
|
|
|
|
|
|
|
A |
- |
1.25 |
1.08 |
10.54 |
|
- |
2 |
1.73 |
9.74 |
|
|
T/U |
2.03 |
- |
20.80 |
0.61 |
|
3.25 |
- |
19.61 |
0.99 |
|
|
C |
2.03 |
24.01 |
- |
0.61 |
|
3.25 |
22.64 |
- |
0.99 |
|
|
G |
34.72 |
1.25 |
1.08 |
- |
|
32.09 |
2 |
1.73 |
- |
|
In Table 4, we provided the ML estimates of transition/transversion bias (R) and respective evolutionary distance values for the GMB haplotypes, their genetically closest cattle individuals, and these two cattle groups combined.
|
Evolutionary characteristics |
Targeted population |
||
|
|
GMB |
GMB genetically closest cattle globally |
Both cattle groups combined |
|
Overall average disparity index (Id) |
0.0023750587 |
0.0012190602 |
0.0014805106 |
|
Overall average composition distance |
0.0072881090 |
0.0048344067 |
0.0054175153 |
|
Average nucleotide composition for T(U)/C/A/G |
25.1/21.8/40.8/12.4 |
25.1/21.7/40.8/12.4 |
25.1/21.7/40.8/12.4 |
|
Nucleotide frequencies (%) respectively for A/T(U)/C/G |
40.76/25.09/21.76/12.39 |
40.78/25.11/21.74/12.37 |
25.1/21.7/40.8/12.4 |
|
ML estimate of transition/transversion bias (R) |
6.23 |
4.01 |
7.69 |
|
Overall average pairwise distance |
0.0064039174 |
0.0048494716 |
0.0052283806 |
|
Overall mean distance |
0.01 |
0.00 |
0.01 |
|
Discrete gamma distribution |
0.0500 |
0.1000 |
0.1000 |
As shown, we only found some differences in the transition/transversion bias between the GMB haplotypes and their genetically closest cattle individuals worldwide: For GMB, the ML estimates (for A↔G and T [U] ↔ C) were collectively slightly higher in contrast to the MCL estimates (for T [U] ↔ C) being slightly lower as compared to these estimates determined for the group of its genetically closest cattle individuals worldwide. Similarly, some slight differences could be also found between the above cattle groups in the MEGA-generated values (Table 3) determined for Id, the overall average composition and pairwise distances, the discrete gamma distribution, and the other parameters including average nucleotide composition, as well as nucleotide frequencies. However, as demonstrated, these values were still almost always greater for the GMB haplotypes as compared to their genetically closest cattle individuals. Furthermore, the average genetic distances within the population of the GMB and its genetically related cattle group were 0.01 and < 0.00, respectively. The calculated average distance between these two groups was 0.00557.
In the analysis of the targeted mtDNA region, using DnaSP, we could identify a 160-bp conserved region (the coordinates according to the B. taurus reference genome [NC_006853.1]: 15629-15788) exhibiting the genetic loci involved in coding for cytochrome b and tRNA-Thr. The other DnaSP-generated statistics are presented in Table 5.
|
Evolutionary characteristics |
Targeted population |
||
|
|
GMB |
GMB closest cattle globally |
Both cattle groups combined |
|
No. of polymorphic sites |
22 |
15 |
26 |
|
Total No. of mutations |
24 |
16 |
28 |
|
Average No. of nucleotide differences (K) |
3.123 |
2.367 |
2.551 |
|
Nucleotide diversity (Pi) |
0.00636 |
0.00482 |
0.00520 |
|
Theta (per site) from Eta |
0.012 |
0.00667 |
0.008 |
|
Invariable (monomorphic) sites |
469 |
476 |
465 |
|
Variable (polymorphic) sites |
22 |
15 |
26 |
|
Singleton variable sites |
13 |
4 |
13 |
|
Parsimony informative sites |
9 |
11 |
13 |
|
Singleton variable sites (2 variants)* |
13 |
4 |
13 |
|
Parsimony informative sites (2 variants)* |
8 |
10 |
12 |
|
Singleton variable sites (3 variants) |
0 |
0 |
0 |
|
Parsimony informative sites (3 variants) |
0 |
1 |
0 |
|
Singleton variable sites (4 variants) |
0 |
0 |
0 |
|
Parsimony informative sites (4 variants) |
1 |
0 |
1 |
|
Sequence conservation (C) |
0.947 |
0.947 |
0.947 |
|
No. of haplotypes (h) |
25 |
20 |
34 |
|
Haplotype diversity (Hd) |
0.997 |
0.930 |
0.950 |
|
Variance of haplotype diversity |
0.00014 |
0.00013 |
0.00007 |
|
Standard deviation of haplotype diversity |
0.012 |
0.011 |
0.008 |
|
DNA conserved region |
13-173 |
13-173 |
13-173 |
As shown, in the DnaSP analysis, we could detect 12 polymorphic mutations across this mtDNA region collectively in the MGB haplotypes, which, in contrast, appeared to be monomorphic in the population of their genetically closest cattle individuals; and vice versa, the DnaSP identified four mutations that were polymorphic in the latter, while exhibiting the monomorphic patterns in the group of GMB haplotypes; a total of 12 mutations were shared by the above two groups; The average number of nucleotide differences (K) versus that of the nucleotide substitutions per site (Dxy) between GMB and the group of its genetically closest cattle individuals were 2.717 and 0.00553 respectively, while the net nucleotide substitutions (Da) per site between these two groups was -0.00006.
Analysis of evolutionary divergence mechanisms
To determine the mechanisms of evolutionary divergence of the GMB haplotypes and their genetically closest cattle individuals, the LD degree across the targeted mtDNA region was assessed. The scatter graphs (A-C), shown in Figure 2, provide the LD values plotted across the nucleotide distance estimates for both the above two groups separately and these groups combined.
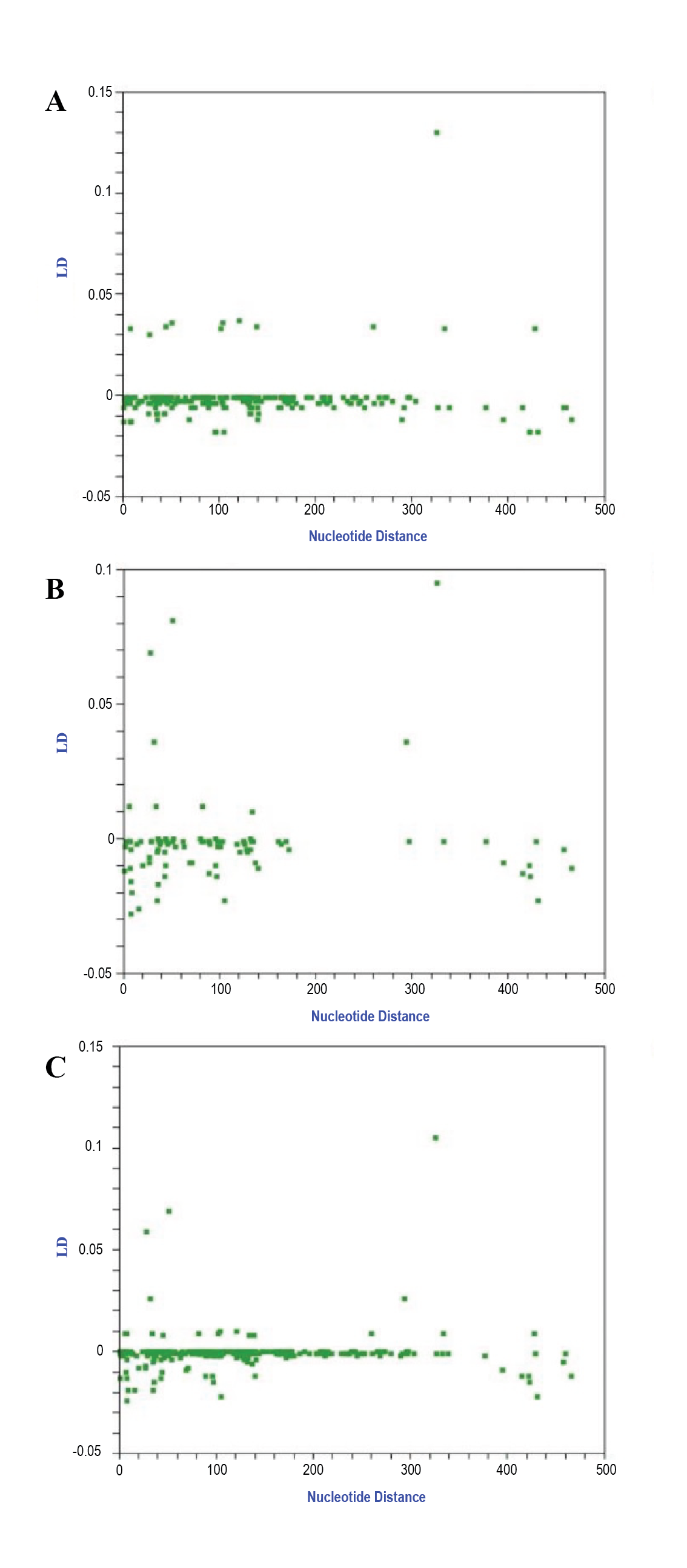
Among the multiple LD-linked polymorphic sites in the mtDNA sequence alignment, different positions were identified across the cattle groups examined: for the GMB haplotypes (n = 26), the 12-338 site positions were highly supported by a strong F-generated p-value (< 0.001) and a Chi-square estimate of 26.000 (p < 0.001) after Bonferroni correction. For the cattle group genetically most closely related to GMB (n = 75), the sites 12-306, 12-338, 407-435 and 427-478 could be determined, strongly supported by the F-produced p-values (< 0.001) and Chi-square estimates (26.172, 75.00, 42.391, 48.340; p < 0.001) after Bonferroni correction. For these two cattle groups combined (n = 101), the interlinked site positions 12-338, 407-435, and 427-478 could be determined and verified by the F-produced p-values (< 0.001) and the robust Chi-sq estimates (101.000, 43.148, 61.137; p < 0.001 respectively) after Bonferroni correction. The DnaSP-generated ZZ values, calculated for inferring intragenic recombination separately between the GMB haplotypes, their genetically most closely related conspecifics, and the total population, were -0.0236, -0.0044, and -0.0157 respectively. In the DnaSP analysis, Rm = 1, with the detected recombined regions located between the following sites for the above cattle groups respectively: 407-435, 427-478 and 389-434. In contrast, in the RDP4 analysis of the targeted mtDNA region, using RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan and 3Seq, we could not detect genetic recombination events that would be supported by statistically reliable values across the above entire population examined in this study.
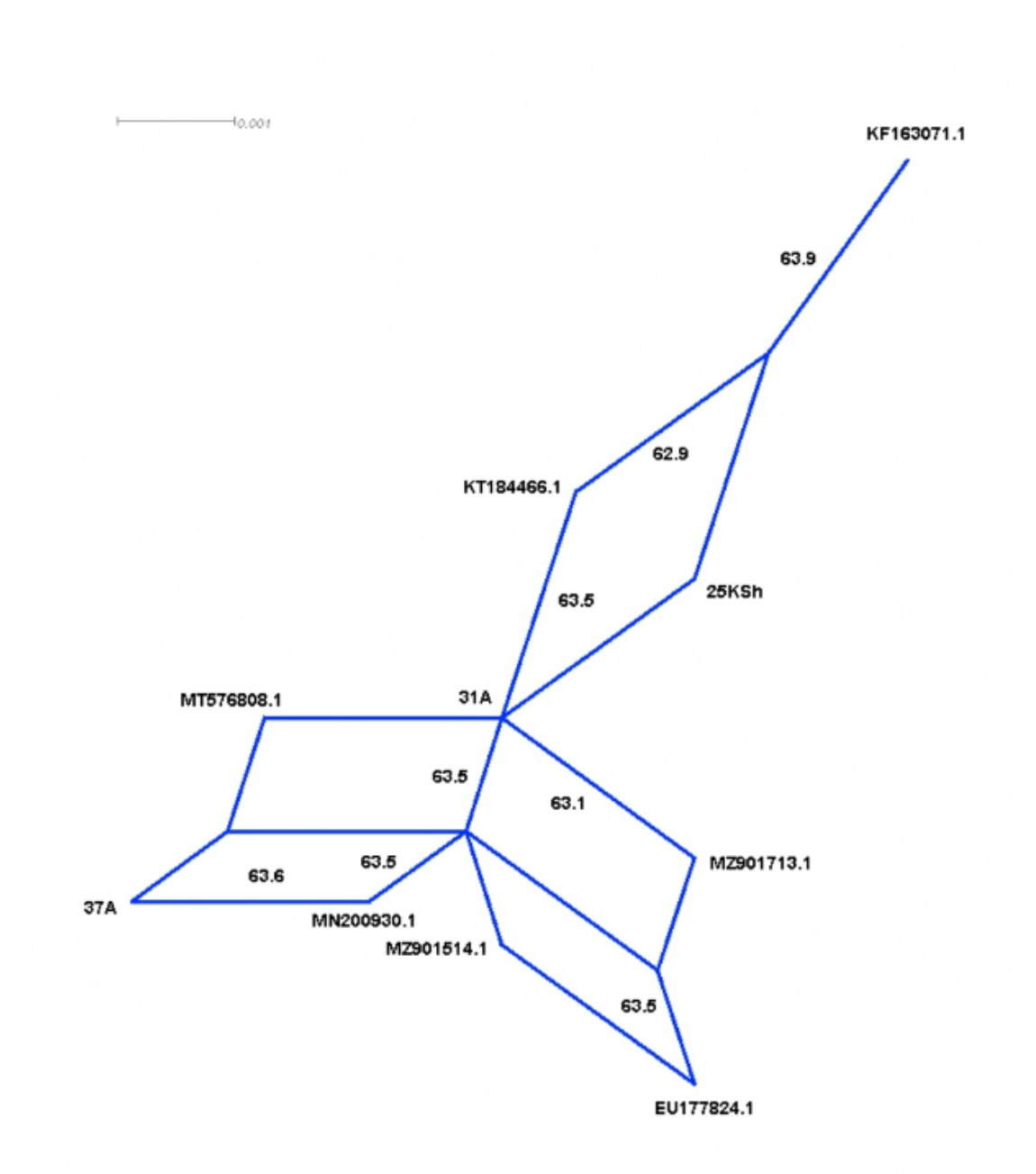
When applying the method of split decomposition, in the SplitsTree analysis of the same mtDNA region, we could determine parallel nucleotide substitutions consolidated into five parallelograms shared by multiple individuals from the populations of the GMB haplotypes and their genetically closest cattle individuals. Importantly, as shown in Figure 3, while the highest fit value of 100 was obtained for the above split decomposition inferences, the bootstrap values, calculated for the nodes of these five parallelograms, were significantly lower, being ≤ 63.6. Moreover, the Phi test, when measuring homoplasy across the targeted mtDNA region for the above subset of cattle individuals, resulted in a very insignificant p-value of 0.4093.
Discussion
Haplotype diversity and population structures of GMB and its genetically closest cattle individuals
Most studies investigating the haplotype diversity and evolution of B. taurus have concentrated on the highly variable D-loop region of mtDNA (Colominas et al., 2015; Kunelauri et al., 2019). Although the D-loop is the most diverse functional region of the mitochondrial genome, several other genetic loci of the mitochondrial genome also show significant polymorphism. Nevertheless, when analyzing the DNA sequences of the D-loop region, some studies have struggled to consistently identify certain haplotypes (e.g. P and T5) in different B. taurus populations globally (Cubric-Curik, 2021). Furthermore, the analysis of the D-loop hypervariable loci has sometimes failed not only in distinguishing between specific breeds but also between some ancient branches (Achilli et al., 2009; Xia et al., 2019). DNA sequencing and analyses of the complete mitogenomes provided new and important insights into the genetic diversity and evolution of B. taurus (Achilli et al., 2008; Cubric-Curik, 2021; Xia et al., 2019; Xia, 2021). However, the cost of DNA sequencing for complete mitogenomes is still not easily affordable for most low- and middle-income countries. In earlier studies, the DNA sequencing of CYTB loci also appeared very instrumental in revealing high haplotype variability (Tarekegn et al., 2018), genetic differences between specific breeds from different countries (Kim, 2013), and even male-mediated introgression (Kikkawa et al., 2003) in B. taurus. In our study, the DNA sequencing and analysis of the D-loop, CYTB, tRNA-Thr, and tRNA-Pro encoding genetic loci could not differentiate, in many instances, between GMB and multiple other breeds from the Turano-Mongolian, European taurine, and some other cattle within the T and Q sub-lineages. Although highlighting the necessity for examining complete mitogenomes, as discussed further, our findings also offer new insights into the haplotype diversity of GMB and the molecular-genetic mechanisms governing the evolution of this breed and its genetically closest cattle individuals.
Interestingly enough, according to the results obtained from our analysis of the targeted mtDNA region, the GMB haplotype diversity was notably greater than previously determined in two pilot studies. However, these pilot studies used a very limited sample size, with the initial investigation including 17 individuals (Kunelauri et al., 2019), and the subsequent one selecting 5 individuals from the original 17 (Kunelauri et al., 2022). Nevertheless, these authors (Kunelauri et al., 2022) reported on several GMB haplotypes falling outside of the known taurine diversity in their analysis of the complete mitogenomes of these cattle individuals. In our study, while the haplotype diversity estimate was comparatively smaller (0.997 versus 0.9995), the Pi value (0.00636) appeared to be notably greater than the mt genome-wide nucleotide diversity estimates (0.0015 and 0.0010-0.0020) calculated earlier respectively for taurine cattle from southeast Europe and some other European regions (Cubric-Curik, 2021). Thus, at least three different possible scenarios can be considered when attempting to explain the observed differences between GMB and the European cattle breeds: as compared with the latter, GMB may have an older evolutionary history, accumulating over time more genetic changes, and/or has evolved at higher evolutionary rates; It is also possible that, compared with GMB, the southeast European breeds might have undergone more extensive selection. These observations could pave a new avenue for future research to better understand the selection-driven evolutionary differences that can potentially exist between these two groups of cattle.
The domestication of B. primigenius, which took place around 10,000 years ago, marked a significant Neolithic advancement (Bonfiglio et al., 2010). This process involved cattle breeding over various periods and had profound socioeconomic impacts on Old World populations (Clutton-Brock, 1989), possibly including early tribes living in Georgia. It's worth noting that historically, dating back 1.8 million years, the territory of Georgia was inhabited by a variant(s) of Homo erectus (Lordkipanidze et al., 2013; Schwartz, Tattersall, & Z, 2014). In this light, it is possible that the above geographic region, being the habitat for certain early tribes, could be one of the oldest cattle domestication sites in the Old World. This scenario gains plausibility given the ancient traditions of winemaking (Ieri et al., 2021), honey production (Kvavadze, Gambashidze, Mindiashvili, & Gogochuri, 2007) and wheat cultivation (Gogniashvili, Matsuoka, & Beridze, 2021) in Georgia. Unfortunately, there is no clear scientific information available about ancient animal husbandry practices and traditions in Georgia.
Here, we show that the GMB Khevsurian and Adjarian populations belong predominantly to the T1, T2, T3 and T4 sub-haplogroups. It should be noted that the investigations, examining complete mitogenomes, revealed significant diversity across modern cattle within T (Cubric-Curik, 2021). The T sub-haplogroups primarily reflect specific geographical structuring, with T1 being most common in African breeds, T2 prevalent in Near Eastern and Mediterranean breeds, T3 predominantly found in European breeds, and T4 most frequently characteristic of East Asian breeds (Carvajal-Carmona et al., 2003; Chen, 2010). The earlier studies reported that, in Europe, T3 demonstrated a reduced diversity relative to Near Eastern cattle – consistent with the patterns found in European cattle in the Near East (Carvajal-Carmona et al., 2003; Troy et al., 2001). In our study, two unique haplotypes, representing the SNGTs, could be identified among the GMB individuals that belonged to T3. Interestingly, T4 could not be found previously in Near Eastern breeds within T (Troy et al., 2001). Here, we show that, similar to the East Asian and some other cow breeds, the GMB Adjarian population also can carry the sub-haplogroup T4 characteristics. According to our phylogenetic analysis, certain GMB SNGTs demonstrated their closest genetic affinities with some sub-linages within T and Q, while some others appeared to represent previously unidentified haplogroups of B. taurus. Importantly, the haplogroup Q, which is likely of Near Eastern origin, has been considered to be rare in the global populations of modern taurine cattle (Bonfiglio et al., 2010; Cubric-Curik, 2021; Xia, 2021). Here, we provide strong amplifying evidence for the growing existence of cattle individuals belonging specifically to Q1 – the sub-lineage of Q – across the GMB populations, strengthening the earlier findings of two pilot studies (Kunelauri et al., 2022; Kunelauri et al., 2019). Moreover, our analysis of the targeted mtDNA region of the GMB populations revealed the possible presence of an unknown sub-haplogroup within Q. Thus, the present findings point out that Georgia should be added to the list of countries (such as Egypt, China, Turkey and several European countries), where the B. taurus populations exhibit Q along with other haplogroups (Achilli et al., 2009).
Among the typical representatives of the GMB Adjarian population, we could identify as well the mtDNA pattern that was most closely related to the sub-haplogroup I1a. I1a is a relatively novel sub-haplogroup of B. indicus (Chen, 2018), which originated in Indus Valley about 8,000 years ago, and further spread eastwards to Southeast Asia and southern China < 400 years ago (Chen, 2010; Loftus, Machugh, Bradley, Sharp, & Cunningham, 1994). Indicine cattle were found to be dominant in southern China (Li, 2013), which is considered to be one of the domestication centres. Interestingly, it is suggested that within I1 cattle found in Guangxi, I1a represents a unique and dominant sub-haplogroup while being absent in India, demonstrating the dominant status for local cattle in Yunnan located in the southwestern part of China (Xia et al., 2019). Thus, similar to earlier observations on cattle (Achilli et al., 2008; Edwards, 2007), the closest genetic affinity of the specific SNGT of GMB with I1a suggests the existence of local admixture populations in this breed, likely influenced by introgression from wild aurochs. Besides, the above highlights can be also collectively suggestive of the complex patterns of the domestication process contributing to shaping the GMB population structure; these findings are in strong agreement with the early studies on the complexity of the domestication process as being a phenomenon influencing the genetic diversity and divergence of cattle populations globally (Achilli et al., 2009; Bonfiglio et al., 2010; Olivieri, 2015; Xia et al., 2019).
Hence, considering all the above findings and observations, we suggest that GMB may have an ancient origin, playing an important role in the evolution of B. taurus globally. More extensive research, using at least the DNA sequencing of complete mitogenomes with significantly larger sample sizes, is needed to gain more in-depth insights into the evolution of GMB.
mtDNA Polymorphisms and mechanisms of evolutionary divergence of GMB
Examining the mtDNA polymorphisms of GMB, we could unravel the presence of multiple hitherto undescribed mutations, clearly exhibiting a transition bias, the phenomenon observed in Mesolithic wild aurochs (B. primigenius) (Edwards, 2007), as well as in some of the ancient and modern bovine populations (Stock et al., 2009). These mutations, identified across 11 sites of the targeted mtDNA region, appeared to be characteristic predominantly to certain GMB SNGTs, distinguishing them from other haplotypes determined previously in the B. taurus global populations. From the evolutionary patterns, it becomes clear that the observed heterogeneity is pronounced primarily across both the SNGT variable sites with two variants, and the parsimony informative sites with two and four variants, being coupled with the above evolutionary estimates including, but not limited to, h, Hd, Id, R and various overall average and mean distances values calculated.
This is the first study offering some important initial insights into the mechanisms of evolutionary divergence of the GMB populations. It should be noted that using the large-scale mitogenome sequence analysis of B. taurus, the first attempts aimed at detecting genetic recombination events in domestic taurine cattle revealed no evidence for this phenomenon in these animals (Cubric-Curik, 2021). While investigating the mechanisms of evolutionary divergence with split decomposition, we identified parallel nucleotide substitutions with some illuminating the conflicting evolutionary signals across the targeted mtDNA region in the populations of both GMB and its genetically closest cattle individuals identified worldwide. While usually such SplitsTree-derived signals can frequently exhibit lateral genetic transfer events, these putative homologous recombination inferences could not be strongly supported by the fit and bootstrap values in the subsequent analysis using the above software package. The scenario of homologous recombination was neither supported by the Phi test estimates when measuring homoplasy for the same mtDNA subset, suggesting, instead, most likely the presence of parallel or convergent evolution in these B. taurus populations. The lowered LD rates, calculated for the targeted mtDNA region of the above populations, could be additionally linked to possible recombination events across the populations of GMB and its genetically closest cattle individuals globally. However, it is important to consider that by lowering LD, homoplasy can sometimes mimic such recombination events (Tibayrenc & Ayala, 2017). Importantly, this and alternatively the other genetic recombination inferences (e.g. Chi-sq and Rm estimates) – the scenario of intragenic recombination – were also rejected when the negative ZZ values were considered in the DnaSP analyses. Such a similar conflicting scenario, depicting the Chi-sq and Rm positive inferences versus the negative ZZ estimates, was described previously, suggesting that at least Rm can be sometimes inflated by parallel mutations not necessarily associated with genetic recombination (Rozas et al., 2001). Interestingly enough, in the earlier study (Groves & Shields, 1997), the CYTB genes of the takin (Budorcas taxicolor) and muskox (Ovibos moschatus) from the family Bovidae were assumed to have been impacted by convergent evolution. Also importantly, the yak/bison mitochondrial transfer, in light of the parallel accumulation of unique mutations of mtDNA, was also suggested (Zeyland et al., 2012). Therefore, our findings conjointly with the above observations strongly lead to the scenario(s) of parallel and/or convergent evolution in the populations of GMB and its genetically closest cattle individuals. These scenarios, versus the scenario of genetic recombination, become even more plausible if we also consider the negative outcomes received from the RDP4 analyses revealing the absence of recombination breakpoints in the mtDNA region of the targeted B. taurus populations.
Conclusions
The majority of GMB individuals from the Khevsureti and Adjara regions of Georgia belong to the sub-haplogroups T1, T2, T3 (including T3119), T4 and Q1. They also exhibit multiple novel haplotypes, largely represented by SNGTs. Some of these SNGTs may belong to currently unidentified sub-haplogroups or even to previously unknown haplogroups of B. taurus. Notably, the haplogroup Q is common in the GMB populations, unlike many other breeds and populations of B. taurus worldwide. Additionally, the Adjarian population of GMB includes the SNGT-9 that is genetically closest to the sub-haplogroup I1a of B. indicus. It is suggested that parallel and/or convergent evolution, along with introgression, have shaped the GMB population structure in these regions of Georgia. Further in-depth research, particularly through the DNA sequencing of complete mitochondrial genomes from a significantly larger number of GMB individuals, is needed to better understand the origin of this breed and its potential role in the evolution of B. taurus global populations.
Supplemental data
Supplemental Table 1. The genetically closest cattle individuals of the Georgian Mountain breed included in the evolutionary analyses.
Supplemental Figure 1. ClustalX-generated multiple DNA sequence alignment utilized in the evolutionary analyses of the Georgian Mountain Breed and its genetically closest cattle individuals.Acknowledgments
This study was supported by the Shota Rustaveli National Science Foundation of Georgia (SRNSFG) (Grant No.: FR-19-21496). The Lugar Center for Public Health Research at the National Center for Disease Control contributed to the DNA sequencing experiments described in this study.
Author contributions
Givi Basiladze and Leila Tabatadze: Conceptualizing the research, and performing the sampling and selection of GMB-specific types of cattle individuals, also contributing to drafting the manuscript; Ekaterine Gabashvili and Mariam Osepashvili: Performing the DNA sequencing and sequence assembly procedures; Marine Murskhvaladze: Performing the nucleotide quality analysis, also contributing to the DNA sequence assembly procedures. Mamuka Kotetishvili: Contributing to the conceptualization of the research, performing evolutionary analyses, and writing the main manuscript text. All the authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that they have no competing interests.